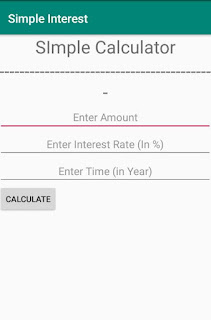
Activity_main.xml File
<?xml version="1.0" encoding="utf-8"?>
<LinearLayout xmlns:android="http://schemas.android.com/apk/res/android"
xmlns:app="http://schemas.android.com/apk/res-auto"
xmlns:tools="http://schemas.android.com/tools"
android:layout_width="match_parent"
android:layout_height="match_parent"
android:orientation="vertical"
tools:context=".MainActivity">
<TextView
android:layout_width="fill_parent"
android:layout_height="wrap_content"
android:text="SImple Calculator"
android:textAlignment="center"
android:textSize="30dp" />
<TextView
android:layout_width="fill_parent"
android:layout_height="wrap_content"
android:text="------------------------------------------"
android:textAlignment="center"
android:textSize="30dp" />
<EditText
android:layout_width="fill_parent"
android:layout_height="wrap_content"
android:hint="Enter Amount"
android:id="@+id/amt"
android:textAlignment="center"
android:inputType="number"/>
<EditText
android:layout_width="fill_parent"
android:layout_height="wrap_content"
android:hint="Enter Interest Rate (In %)"
android:id="@+id/interest"
android:textAlignment="center"
android:inputType="number"/>
<EditText
android:layout_width="fill_parent"
android:layout_height="wrap_content"
android:hint="Enter Time (in Year)"
android:id="@+id/tim"
android:textAlignment="center"
android:inputType="number" />
<Button
android:layout_width="wrap_content"
android:layout_height="wrap_content"
android:text="Calculate"
android:id="@+id/btn" />
<TextView
android:layout_width="fill_parent"
android:layout_height="wrap_content"
android:id="@+id/txt1"
android:textSize="30dp" />
<TextView
android:layout_width="fill_parent"
android:layout_height="wrap_content"
android:id="@+id/txt2"
android:textSize="20dp" />
</LinearLayout>
MainActivity.Java File
package com.irawen.simpleinterest;
import android.support.v7.app.AppCompatActivity;
import android.os.Bundle;
import android.view.View;
import android.widget.Button;
import android.widget.EditText;
import android.widget.TextView;
public class MainActivity extends AppCompatActivity {
@Override protected void onCreate(Bundle savedInstanceState) {
super.onCreate(savedInstanceState);
setContentView(R.layout.activity_main);
OnCLick();
}
EditText amt,interest,time;
Button btn;
TextView txt1,txt2;
public void OnCLick()
{
amt=(EditText)findViewById(R.id.amt);
interest=(EditText)findViewById(R.id.interest);
time=(EditText)findViewById(R.id.tim);
btn=(Button)findViewById(R.id.btn);
txt1=(TextView)findViewById(R.id.txt1);
txt2=(TextView)findViewById(R.id.txt2);
btn.setOnClickListener(new View.OnClickListener() {
@Override public void onClick(View v) {
int a=Integer.parseInt(amt.getText().toString());
int b=Integer.parseInt(interest.getText().toString());
int c=Integer.parseInt(time.getText().toString());
int d;
d=(a*b*c)/100;
int e=a+d;
txt1.setText("Total Interest Is :"+String.valueOf(d));
txt2.setText("Total Amount is : "+String.valueOf(e));
}
});
}
}














.png)






















.png)






